About me
Hello! I’m Aaron, a PhD candidate at the University of California San Diego in the Center for Marine Biotechnology and Biomedicine at Scripps Institution of Oceanography. I have worked in the lab of Scripps Department Chair Prof. Eric Allen since 2018. My research focuses on in silico analyses of marine microbial communities, with a specialization in the genomics of algae and their microbiomes.
Before starting my PhD, I got my bachelor’s degree in Bioinformatics at UC San Diego. I worked as an instructional assistant for two years at the Jacobs School of Engineering during undergrad. My specialty was the theoretical bases of computer science, with a focus on discrete mathematics, computational complexity, and algorithm design. I leverage my background in computer science to explore communities of marine microorganisms for potential applications in biotechnology, bioenergy, and biomaterials.
Research Aims
I love collaboration, so my time is split between many projects! My dissertation is centered around the strategies used by microbes to degrade algal polysaccharides and defense chemicals. These enzymes and compounds have implications in aquaculture, carbon sequestration, and marine ecosystem dynamics. Parallel to my disseration, I partner with corporate R&D teams, government labs, and academic research groups as both a generalist bioinformatician and a microbial genomics specialist.
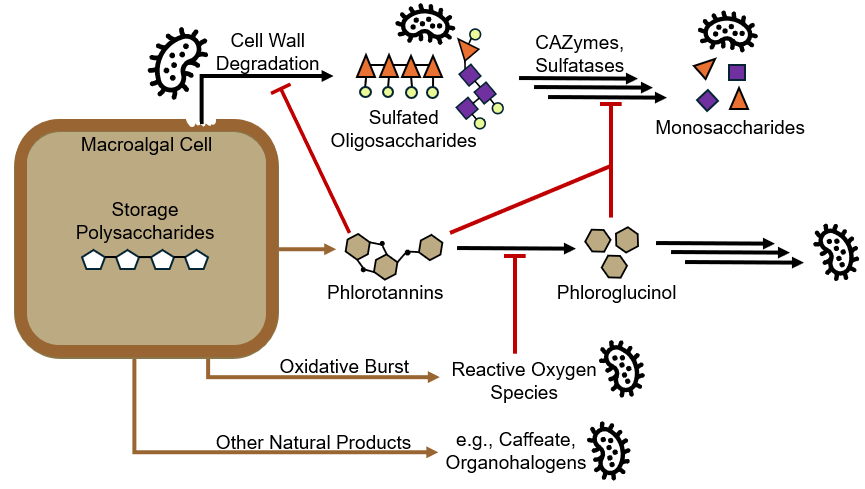 |
|---|
| My study system: the defensive cascade of interactions between algal cells and their microbiomes |
Microbiome decomposition of macroalgal polysaccharides and antimicrobials
My dissertation work focuses on the sugars inside seaweeds, which are notoriously difficult to break down using physical or chemical treatments. I use the gut of the herbivorous Hawaiian chub as a model system for studying the bacteria that simultaneous degrade wide varieties of algal sugars and detoxify the antimicrobial compounds released by stressed seaweeds. These microbial communities can survive for multiple weeks in batch-fed bioreactors (see my dissertation chapter), enabling direct manipulation of seaweed-degrading microbiomes. Using this paired natural fish/bioreactor model system, my research provides a comprehensive view of the temporal dynamics, microbiogeography, genome organization, and enzyme evolution behind seaweed biomass consumption.
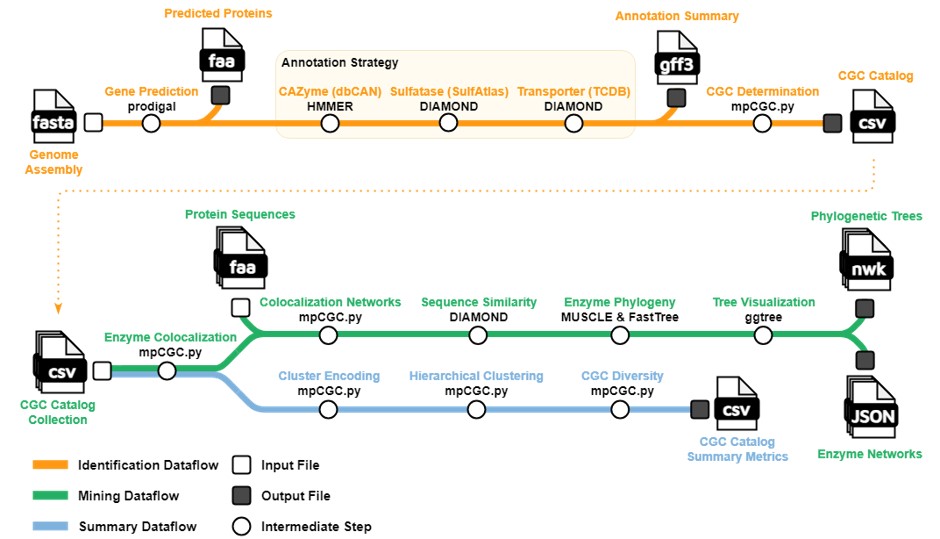 |
|---|
| Dissertation work: a pipeline for identifying macroalgal polysaccharide degradation gene clusters |
Microalgal genomics and genetic engineering toolkits
I am also interested in developing genomic resources and genetic engineering platforms for commercially-relevant green microalgae strains. I collaborate with molecular biologists at the California Center for Algal Biotechnology and the J. Craig Venter Institute to catalog resillience of novel Chlamydomonas strains. Some strains display promising tolerance to temperature, pH, salt, and light conditions that limit the growth of similar algae used by algaculture farms. As a bioinformatician in one branch of this project, I harness information encoded in their assembled genomes to identify genes that enable this extremophile tolerance. I also develop computational tools for genetic engineering of these algae strains, which enable 10-fold improvements in lipid production and microalgae that are able to degrade polyethylene terephthalate microplastics.
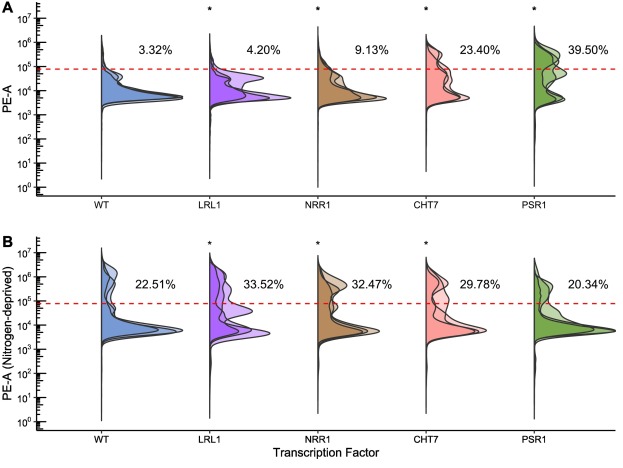 |
|---|
| Collaboration work: improving lipid accumulation in genetically engineered green microalgae |
Sustainable biotechnology applications
I often partner with Californian and Hawaiian biotechnology companies to extend the impact of my research. Much of my disseration work is done in tandem with applied scientists at the Kona mariculture firm Ocean Era, to use partially degraded macroalgal biomass as a sustainable feed for carnivorous fish farms. My bioinformatics support work in green microalgae enables automated pathogen monitoring systems for the outdoor commercial ponds of Hawaiian algaculture company Global Algae.
More recently, I have built computational platforms for genetic manipulation of a proprietary algae strain used by Algenesis, a local biomaterials startup that produces 100% biodegradable algae-based plastics. Current, I work with BASF’s enzyme division on an ongoing grant from the Department of Energy to halve the carbon footprint of industrial surfactant production using marine enzymes indentified in my dissertation.
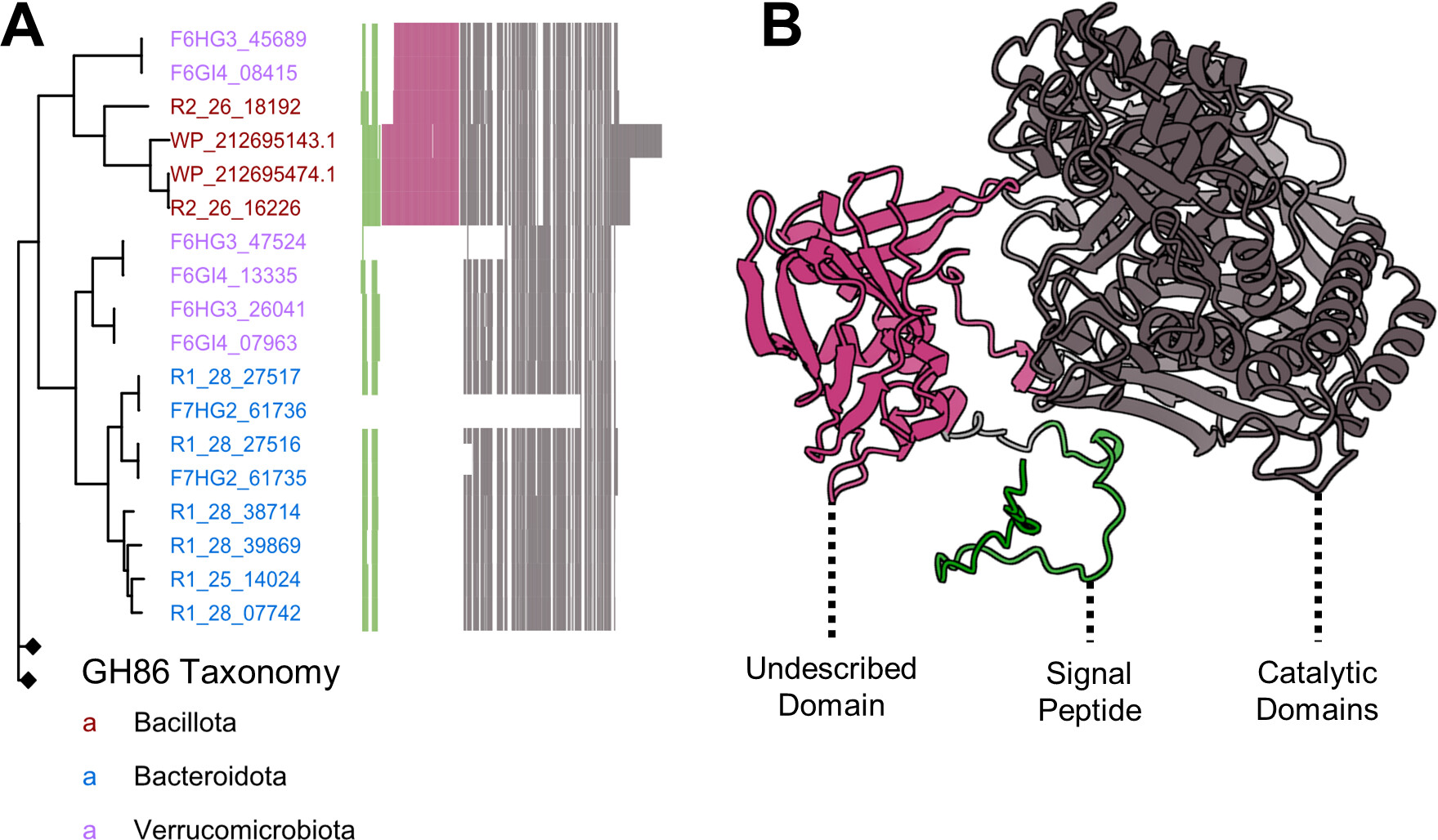 |
|---|
| Public-private partnership: heterologous expression of novel marine enzymes (e.g. β-porphyranase) for biofoundries |
Microbial ‘omics across environments
Outside of my primary research projects, I also collaborate with wet lab biologists to integrate computational approaches into their studies. These contributions often extend beyond my primary niches of algal-associated genomics. These projects include investigating the metabolic capacity of sponge microbiomes, the infection strategies of deep sea viruses, elephant pathogens in Botswana, and the responses of a keystone crustacean to stresses associated with climate change.
